|
|
|
|||||||||||||||||||
|
||||||||||||||||||||||
|
We study how neural circuits are organized to process information, and how they are assembled during development. To address these questions, we use fruit flies and mice as model organisms, and combine advanced molecular genetics with anatomical, physiological and behavioral approaches. Organization of the olfactory system 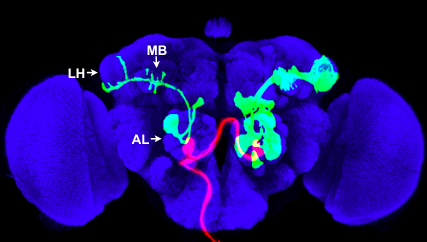 |
| Fig. 1: Red: axon projections of a single ORN class from the left maxillary palp, which terminate in a pair of glomeruli in the antennal lobe (AL). Green: a single PN (left) and a group of PNs that share the same lineage (right) labeled by MARCM; they project dendrites into the antennal lobe, and axons to the mushroom body (MB) and lateral horn (LH). Blue: neuropil marker. |
|
The olfactory systems from flies to mammals use a similar organizational principle. In the fly, olfactory receptor neurons (ORNs) expressing the same odorant receptor project their axons to the same glomerulus in the antennal lobe. Projection neurons (PNs) send dendrites to individual glomeruli, and relay olfactory information via their axons to the mushroom body and lateral horn (Fig. 1). Using MARCM (see below) to label individual neurons, we previously found that PN axon terminals exhibit striking stereotypy at the lateral horn according to the glomeruli they send dendrites to. Axon terminals of PNs representing food odors are spatially segregated from those that represent mating pheromones. By contrast, PN axon terminal arborizations in the mushroom body, the olfactory learning and memory center, exhibit much less stereotypy (Jefferis et al., 2007; Marin et al., 2002). Recently, we have also characterized arborization patterns of local interneurons in the antennal lobe, and found considerable diversity and variability in their wiring patterns (Chou et al., 2010). We are currently using two-photon calcium imaging, optogenetics, and quantitative behavioral assays to identify principles of information processing at the antennal lobe and in higher olfactory centers suggested by previous anatomical studies. Analogous to flies, mouse ORNs expressing the same odorant receptor project their axons to the same glomerulus in the olfactory bulb, creating a spatial map for odor processing. Little is known about how the olfactory bulb glomerular map is represented in the olfactory cortex. We are currently investigating this problem using virus-mediated trans-synaptic tracing (Miyamichi et al., unpublished). Development of wiring specificity in the fly olfactory system The assembly of the fly olfactory system requires precise glomerular targeting of axons from each of the 50 ORN classes, as well as dendrites of each of the 50 PN classes (Jefferis et al., 2001). We are using this neural circuit as a model to investigate the general principles by which precise wiring specificity arises during development. Our previous studies have shown that PN dendrite patterning precedes ORN axon targeting (Jefferis et al., 2004). PN dendrite targeting relies on global cues in the form of gradients (Komiyama et al., 2007), as well as local cues distributed in a “salt-and-pepper” fashion on dendrites projecting to different glomeruli (Hong et al., 2009). Targeting of ORN axons may use the same molecules as PN dendrite targeting, but via distinct mechanisms including axon-axon interactions (Sweeney et al., 2007) and axon-target interactions (Chou et al., 2010). We are currently performing systematic genetic screens coupled with targeted candidate gene approaches in an effort to identify the cell-surface code ORNs and PNs use to form specific connections at stereotypically organized glomeruli. Developmental neurobiology In addition to our focus on the olfactory system, we are studying several other developmental neurobiological problems. For example, we have shown that fly mushroom body γ neuron axons undergo stereotypical developmental pruning. This pruning process utilizes a local axon degeneration mechanism, and requires cell autonomous action of a steroid hormone ecdysone receptor and the ubiquitin-proteasome system, as well as glial engulfment receptors that mediate removal of degenerated axons (Hoopfer et al., 2006; Lee et al., 2000; Watts et al., 2003). We are currently studying cell-cell communications that regulate axon pruning. Recently, using a genetic mosaic system we have developed in the mouse (see below), we have uncovered novel roles for genes important for neuronal migration (Hippenmeyer et al., 2010). In addition, we have identified a cell-autonomous role for the NMDA receptor in patterning dendrites according to input organization (Espinosa et al., 2009). We continue to investigate genes that regulate neuronal activity for their function in the maturation of individual neurons and their integration into functional circuits. Creating genetic tools 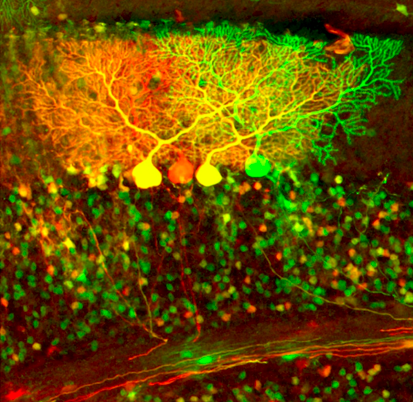 Fig. 2: Cerebellar neurons labeled by MADM In the process of dissecting the adult organization and developmental assembly of complex neural circuits, we have created several useful genetic tools. The MARCM method (Mosaic Analysis with a Repressible Cell Marker) (Lee and Luo, 1999) enables the visualization and genetic manipulation of small populations of cells or single neurons in a mosaic fly (Fig. 1). It has been instrumental in many of the projects described above and has been widely used in the Drosophila field. Recently we have developed a new repressible binary expression system, the Q system (Potter et al., 2010). The Q system has many applications, and we are applying it to several problems described above. We have also developed a mosaic method in the mouse called MADM (Mosaic Analysis with Double Markers) (Zong et al., 2005), which is conceptually similar to fly MARCM. MADM allows sparse labeling and genetic manipulation of individual cells or cells that share the same lineage with distinct colors in mosaic animals (Fig. 2). Using MADM as a lineage analysis tool, we have shown that cerebellar granule cells that share a common lineage project their axons to a specific sublayer of the molecular layer (Espinosa and Luo, 2008). We are currently expanding the MADM technique to other mouse chromosomes, and will use MADM to explore neural developmental processes (see above). We have also developed other useful mice, such as a double fluorescent Cre reporter (Muzumdar et al., 2007) and synapse labeling tools in vivo (Li et al., 2010). Chou, Y.H., Spletter, M.L., Yaksi, E., Leong, J.C., Wilson, R.I., and Luo, L. (2010). Diversity and wiring variability of olfactory local interneurons in the Drosophila antennal lobe. Nat Neurosci 13, 439-449. Chou Y.H., Zheng X., Beachy PA, Luo L (2010) Patterning axon targeting of olfactory receptor neurons by coupled Hedgehog signaling at two distinct steps. Cell 142, 954-966. Espinosa, J.S., and Luo, L. (2008). Timing neurogenesis and differentiation: insights from quantitative clonal analyses of cerebellar granule cells. J Neurosci 28, 2301-2312. Espinosa, J.S., Wheeler, D.G., Tsien, R.W., and Luo, L. (2009). Uncoupling dendrite growth and patterning: single-cell knockout analysis of NMDA receptor 2B. Neuron 62, 205-217. Hippenmeyer S, Youn YH, Moon HM, Miyamichi K, Zong H, Wynshaw-Boris A, Luo L (2010) Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron in press. Hong, W., Zhu, H., Potter, C.J., Barsh, G., Kurusu, M., Zinn, K., and Luo, L. (2009). Leucine-rich repeat transmembrane proteins instruct discrete dendrite targeting in an olfactory map. Nat Neurosci 12, 1542-1550. Hoopfer, E.D., McLaughlin, T., Watts, R.J., Schuldiner, O., O'Leary, D.D., and Luo, L. (2006). Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron 50, 883-895. Jefferis, G.S., Potter, C.J., Chan, A.M., Marin, E.C., Rohlfing, T., Maurer, C.R., Jr., and Luo, L. (2007). Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell 128, 1187-1203. Jefferis, G.S., Vyas, R.M., Berdnik, D., Ramaekers, A., Stocker, R.F., Tanaka, N.K., Ito, K., and Luo, L. (2004). Developmental origin of wiring specificity in the olfactory system of Drosophila. Development 131, 117-130. Jefferis, G.S.X.E., Marin, E.C., Stocker, R.F., and Luo, L. (2001). Target neuron prespecification in the olfactory map of Drosophila. Nature 414, 204-208. Komiyama, T., Sweeney, L.B., Schuldiner, O., Garcia, K.C., and Luo, L. (2007). Graded expression of semaphorin-1a cell-autonomously directs dendritic targeting of olfactory projection neurons. Cell 128, 399-410. Lee, T., and Luo, L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451-461. Lee, T., Marticke, S., Sung, C., Robinow, S., and Luo, L. (2000). Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron 28, 807-818. Li, L., Tasic, B., Micheva, K.D., Ivanov, V.M., Spletter, M.L., Smith, S.J., and Luo, L. (2010). Visualizing the distribution of synapses from individual neurons in the mouse brain. PLoS One 5, e11503. Marin, E.C., Jefferis, G.S.X.E., Komiyama, T., Zhu, H., and Luo, L. (2002). Representation of the glomerular olfactory map in the Drosophila brain. Cell 109, 243-255. Muzumdar, M.D., Tasic, B., Miyamichi, K., Li, L., and Luo, L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605. Potter, C.J., Tasic, B., Russler, E.V., Liang, L., and Luo, L. (2010). The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141, 536-548. Sweeney, L.B., Couto, A., Chou, Y.H., Berdnik, D., Dickson, B.J., Luo, L., and Komiyama, T. (2007). Temporal target restriction of olfactory receptor neurons by Semaphorin-1a/PlexinA-mediated axon-axon interactions. Neuron 53, 185-200. Watts, R.J., Hoopfer, E.D., and Luo, L. (2003). Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 38, 871-885. Zong, H., Espinosa, J.S., Su, H.H., Muzumdar, M.D., and Luo, L. (2005). Mosaic analysis with double markers in mice. Cell 121, 479-492. |
| Updated 09/2010 |